2 File Formats
To facilitate high throughput data manipulation and reduce storage usage, several file format have been proposed and generaly accepted as the standard. Due to these great efforts (e.g. SAM/BAM and VCF), data analysis and tool development become more easier and highly efficient. However, when it comes to bisulfite sequencing data, currently, available tools possess their own tool specific data format. In consequence, integrating results from several tools leads to extra efforts in unifying data format and developing custermized tools, which is time comsuming and error prone.
The widely-used BS-seq alignment software BS-Seeker2 defines CGmap and ATCGmap file formats for the representation of DNA methylomes. In CGmapTools, we used ATCGmap and CGmap as the standard file format interface, so that to simplify the development of downstream DNA methylation analysis tools and to provide standard formats for storing and sharing the DNA methylomes.
In CGmapTools, we designed novel binary formats: CGbz and ATCGbz for less coverage and improvements in random-accessing data in large data in hard-disk.
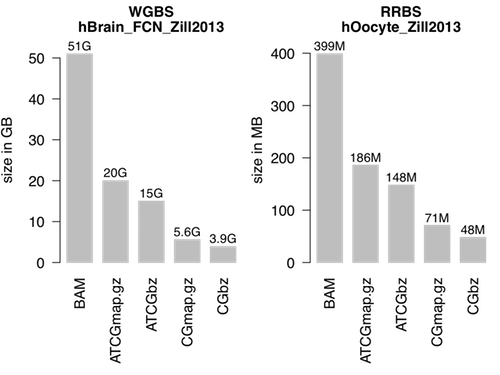
Figure 2.1: Size of multiple file formats
2.1 ATCGmap Format
Similar with pileup, ATCGmap format summarizes the information of mapped reads covered on each nucleotide on both strands, specially designed for BS-seq data.
Here, we defined ATCGmap file format to integrate both mapping and coverage of non-cytosine and cytosine sites with estimated DNA methylation in a single file.
Example
chr1 T 3009410 -- -- 0 10 0 0 0 0 3 0 0 0 na chr1 C 3009411 CHH CC 0 10 0 0 0 0 4 0 0 0 0.0 chr1 C 3009412 CHG CC 0 10 0 0 0 0 9 1 0 0 0.0 chr1 C 3009413 CG CG 0 10 50 0 0 0 20 1 0 0 0.83Column Description

Figure 2.2: Description of ATCGmap
2.2 CGmap Format
In cases we only want to retain DNA methylation on cytonsines to save storage usage, we defined another file format called CGmap which provides sequence context and estimated DNA methylation level of any covered cytosines on the reference genome.
Example
chr1 G 3000851 CHH CC 0.1 1 10 chr1 C 3001624 CHG CA 0.0 0 9 chr1 C 3001631 CG CG 1.0 5 5 chr1 G 3001632 CG CG 0.9 9 10Column Description

Figure 2.3: Description of CGmap
2.3 ATCGbz Format
ATCGbz format is the binary compressed version for ATCGmap format. ATCGmap format is readable, while quite large for storing, and difficult for fetching information in a specific position. ATCGbz is defined as the sorted binary version, that storing all information of ATCGmap into standard binary form, largely reduced the storage requirement, and also supporting fast retrival of methylation information for any position on genome.
- Data structure
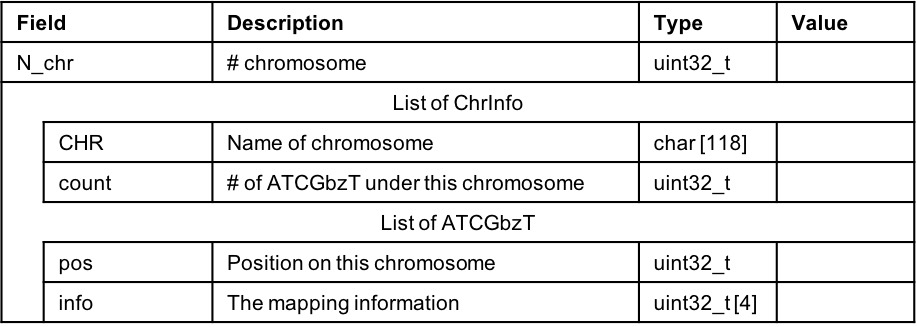
Figure 2.4: Data structure of ATCGbz
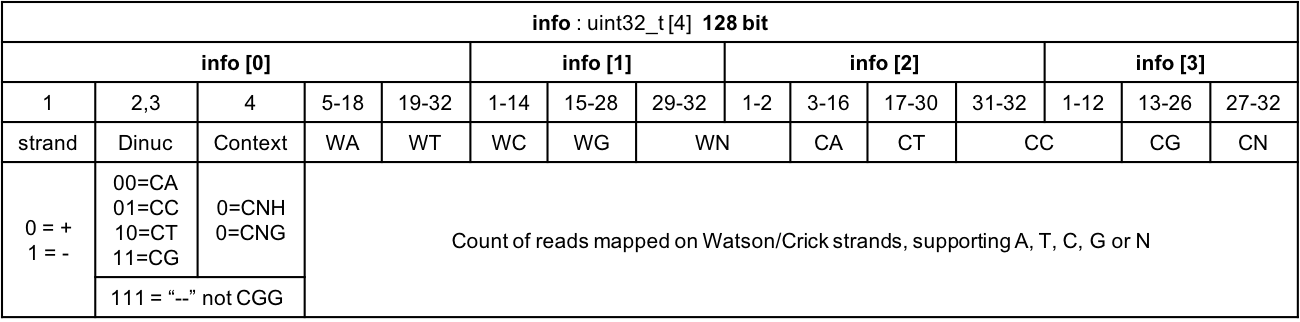
Figure 2.5: Data structure of info field of ATCGbz
- Related command
Command
cgmaptools fetch atcgbz -h #
# Usage: cgmaptools fetch atcgbz -b <ATCGbz> -C <CHR> -L <LeftPos> -R <RightPos>
# (aka ATCGbzFetchRegion)
# Description: Convert ATCGbz format to ATCGmap format.
# Contact: Guo, Weilong; guoweilong@126.com
# Last update: 2016-12-07
#
# Options:
#
# -h, --help output help information
# -b, --ATCGbz <arg> output ATCGbz file
# -C, --CHR <arg> specify the chromosome name
# -L, --leftPos <arg> the left position
# -R, --rightPos <arg> the right position2.4 CGbz Format
CGbz format is the binary compressed version for CGmap format.
- Data structure
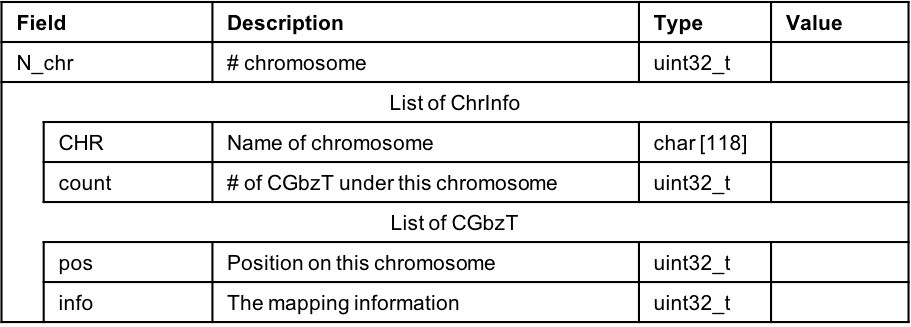
Figure 2.6: Data structure of ATCGbz
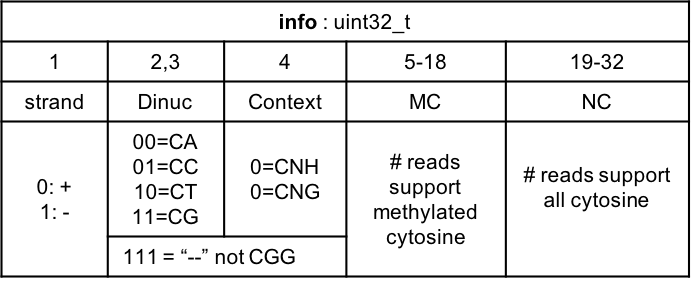
Figure 2.7: Data structure of info field of CGbz
- Related command
Command
cgmaptools fetch cgbz -h #
# Usage: cgmaptools fetch cgbz -b <CGbz> -C <CHR> -L <LeftPos> -R <RightPos>
# (aka CGvzFetchRegion)
# Description: Convert CGbz file to CGmap format.
# Contact: Guo, Weilong; guoweilong@126.com
# Last update: 2016-12-07
#
# Options:
#
# -h, --help output help information
# -b, --CGbz <arg> output CGbz file
# -C, --CHR <arg> specify the chromosome name
# -L, --leftPos <arg> the left position
# -R, --rightPos <arg> the right position